The Biomechanics and Mechano-biology research program led by Professor Sachin Goyal focuses on fundamental research in core Mechanical Engineering areas of Mechanics, Dynamics or Controls motivated by applications in the fields of Biology and Medicine. The primary emphasis is on physical, conceptual and mathematical modeling along with computational simulations, and the applications range from human biomechanics to structural dynamics of micro-scale biological filaments.
Example Research Directions:
1. Modeling Biomechanical Symptoms of Parkinson's Disease
Patients suffering from Parkinson's disease (PD) experience several biomechanical symptoms such as tremor, postural stooping, gait problems and imbalance with tendency to fall. Parkinson's 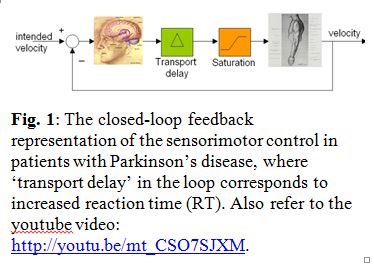 disease is also characterized by a permanent increase in reaction time (RT) in both voluntary and involuntary motor responses. For a healthy person, the RT is about 250 milliseconds, while for a PD patient already experiencing tremor, it’s about 350 milliseconds. Although the deterioration in biomechanical control can intuitively be related to sluggish reaction times, how the increase in reaction time leads to such biomechanical symptoms as tremor and stooping is little known. Recently, we proposed a simple perspective based on delayed-feedback control of mechanical systems, as depicted in Figure on the right, to explain the connection between RT and the biomechanical symptoms. The perspective explains how the cause of the Parkinsonian tremor actually lies in the patient's intent to stop it, and provides grounds for a differential diagnosis of Parkinsonian tremor from other physiological or essential tremors.
disease is also characterized by a permanent increase in reaction time (RT) in both voluntary and involuntary motor responses. For a healthy person, the RT is about 250 milliseconds, while for a PD patient already experiencing tremor, it’s about 350 milliseconds. Although the deterioration in biomechanical control can intuitively be related to sluggish reaction times, how the increase in reaction time leads to such biomechanical symptoms as tremor and stooping is little known. Recently, we proposed a simple perspective based on delayed-feedback control of mechanical systems, as depicted in Figure on the right, to explain the connection between RT and the biomechanical symptoms. The perspective explains how the cause of the Parkinsonian tremor actually lies in the patient's intent to stop it, and provides grounds for a differential diagnosis of Parkinsonian tremor from other physiological or essential tremors.
The above perspective opens up several research opportunities towards understanding the biomechanical aspects of the disease that must transform the medical science behind early diagnosis, disease monitoring and treatment strategies. Below are a few examples:
Smartphone App for Diagnosis and Disease Monitoring
So far, there exists no diagnostic test that can confirm Parkinson’s disease, although computer tomography (CT) and magnetic resonance images (MRI) of the brain, which usually appear normal in PD patients, are sometimes used to rule out other disorders that could give rise to similar symptoms. However, the Parkinsonian tremor, which as per the above perspective would be a limit-cycle oscillation due to delay-induced instability, has rich information for diagnosis that is yet unexploited in medical research. This lends itself into the idea of developing a smartphone application that can analyze tremor and related characteristics and quantify the state and progression of Parkinson’s disease using RT.
Multi-body Dynamics (MBD) based Simulation Model for Posture and Gait Control
Active neural feedback control is essential for humans to stand upright. This is obvious because a dead body cannot maintain an upright position. Sluggish reaction time (RT) in the above perspective tends to destabilize the control of upright posture resulting in a postural compromise that must lead to stooping. Simulations can provide quantitative insights on how the RT affects both the posture as well as gait. The simplest modeling efforts for simulating the dynamics of an upright posture can begin with an inverted pendulum. We are leveraging MBD software to develop a representation of human body with joints at a necessary level of sophistication for gait and posture simulations and a feedback neural controller using MATLAB Simulink or an equivalent tool.
2. Modeling Constitutive Laws of Biological Filaments from their Atomistic Structures
Bending and twisting of biological filaments such as DNA (Deoxyribonucleic acid) are crucial to their biological functions. For example, as shown in the Figure on the right, activity of the genes in lac-operon is 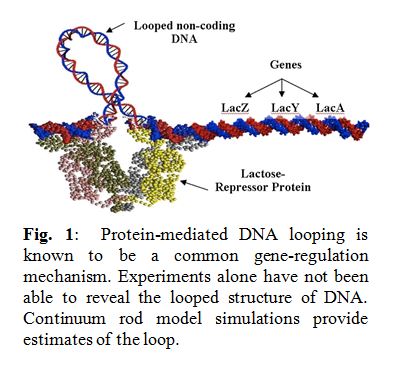 governed by the sequence-dependent looping behavior of its “non-coding” DNA segment (portion of DNA that does not contain genes). Understanding the nonlinear dynamics of these mechanical deformations and how they are caused within the intra-cellular environment provides for a promising foundation for futuristic medical inventions. In fact, this ambitious and scientifically rich research quest is best introduced by posing three fundamental questions:
governed by the sequence-dependent looping behavior of its “non-coding” DNA segment (portion of DNA that does not contain genes). Understanding the nonlinear dynamics of these mechanical deformations and how they are caused within the intra-cellular environment provides for a promising foundation for futuristic medical inventions. In fact, this ambitious and scientifically rich research quest is best introduced by posing three fundamental questions:
- How do the structural deformations of non-coding DNA regulate gene expression?
- How do the gene-regulating proteins manipulate structural deformations in DNA?
- How does base-pair sequence (or the atomistic structure) of non-coding DNA influence its structural deformability?
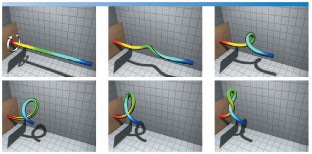 |
| Fig. Torsional buckling of a filament simulated from our computational rod model. For the video animation, refer to the youtube video: https://youtu.be/bGAQ-5wBYiQ. |
While several experimental biophysicists are engaged in answering the first question, our program is addressing the other two questions that ensued a rigorous and basic research in field of mechanics. Recognizing DNA as a thin filament with bending and torsional stiffness, we developed a “rod” model to simulate the biologically relevant looping and intertwining of the DNA molecules. Figure on the right shows an example simulation that involves nonlinear dynamics of torsional buckling with self-contact and intertwining. The model requires solving constrained partial differential equations that pose numerical challenges. It depicts the torsional buckling of a filament simulated from our computational rod model.
There is however a very little effort towards the third question, which actually has the grassroots of the first two questions. We are investigating the third question by analyzing how the atomic configurations and interatomic interactions map into constitutive behaviors at continuum level, i.e. into the restoring effects or “springiness” of the filament in bending and twisting. Since a first-principles derivation of the constitutive law from an atomistic-level structure and interactions is often impractical and so are direct experimental measurements due to the small length-scales, we are developing an “inverse rod model” to estimate the constitutive law from high-fidelity discrete-structure simulations such as molecular-dynamics (MD) simulations.
This research is being led by Dr. Sachin Goyal's Group.



